
Blog post by Noreen Brenner, January 1, 2025
Ancient retroviruses in the vertebrate genome played a role in the evolution of vertebrate brains

Very interesting! Researchers have discovered that ancient retroviruses in the DNA of vertebrates played a role in the evolution of vertebrate brains.
Click on the link below, to learn more:
Ancient retroviruses played a key role in the evolution of vertebrate brains, suggest researchers
**************************************

Blog post by Noreen Brenner, December 26, 2024
Microbiome research has shown that we need to revise our view of microbes as merely pathogens
Click on the link below, to learn more:
https://phys.org/news/2024-03-lexicon-age-microbiome.html
**************************************


Greenland shark at Admiralty Inlet, Nunavut1

“Activation of p53 in response to stress signals initiates its transcriptional activity, leading to the activation of cellular protective pathways – p53 binds to the DNA […] and promotes the transcription of a wide array of genes.”2
************
************
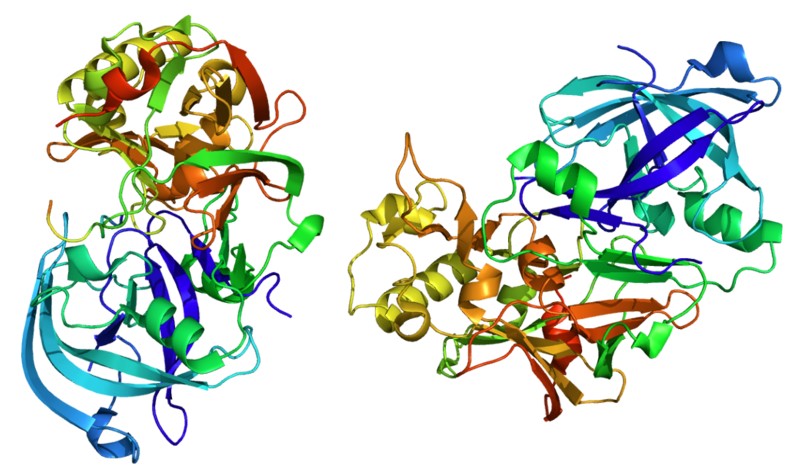
Image attribution: Emw, CC BY-SA 3.0 https://creativecommons.org/licenses/by-sa/3.0, via Wikimedia Commons
-
The genome of the world’s longest-lived vertebrate, the Greenland shark, has been sequenced!
by Noreen Brenner, CEO and Chief Scientific Officer
December 15, 2024
Hooray, the Greenland shark’s genome has been sequenced! It is the longest-lived vertebrate, surviving for around 400 years.
The international group of researchers who sequenced the DNA, found that the genome is enormous due to transposable elements (repetitive stretches of DNA that can multiply). Notably, it seems that genes involved in DNA repair (which confer a longevity benefit to the organism) have used the duplication machinery of the transposable elements to replicate.
In addition, the p53 gene (the guardian of the genome, the most important tumor suppressor gene), which protects against cancer, has modifications which require further study to see how they might be pro-longevity.References:
**************************************

The marvel that is Alphafold, a revolutionary new tool for drug discovery
By Noreen Brenner, CEO and CSO
March 8, 2024
For sixty years, scientists tried to predict protein structure based on protein sequence. This seemed an impossible task, until Google’s Alphafold, an AI, achieved the impossible last year.
From 1994 until 2023, scientists participated every two years in the Critical Assessment of Techniques for Protein Structure Prediction (CASP) competition. CASP’s task was to predict protein structure based on amino acid sequence, without knowing the experimentally determined structure. In September 2023, Alphafold far surpassed all other participants in very accurately predicting protein structure, and not only won the competition but achieved a Holy Grail of biological science.
How did Alphafold manage to win? It used deep learning machine learning algorithms, and was trained on innumerable known protein sequences matched to their known structures (available in the Protein Data Bank or PDB).
Alphafold’s great feat of cracking the code of how amino acid sequences fold to form 3-dimensional protein structures will facilitate drug discovery, as it is a useful tool in identifying targets.
References:
- 60 Years in the Making: AlphaFold’s Historical Breakthrough in Protein Structure Prediction | From Atoms To Words (arturorobertazzi.it)
- CASP14: what Google DeepMind’s AlphaFold 2 really achieved, and what it means for protein folding, biology and bioinformatics | Oxford Protein Informatics Group (blopig.com)
- How DeepMind’s protein-folding breakthrough could transform drug development (thenextweb.com)
- Image attribution: Hemming1952, CC BY-SA 4.0 https://creativecommons.org/licenses/by-sa/4.0, via Wikimedia Commons ↩︎
- Image attribution: Ana Janic, Etna Abad & Ivano Amelio, CC BY 4.0 https://creativecommons.org/licenses/by/4.0, via Wikimedia Commons ↩︎